publications
publications by categories in reversed chronological order. generated by jekyll-scholar.
2024
-
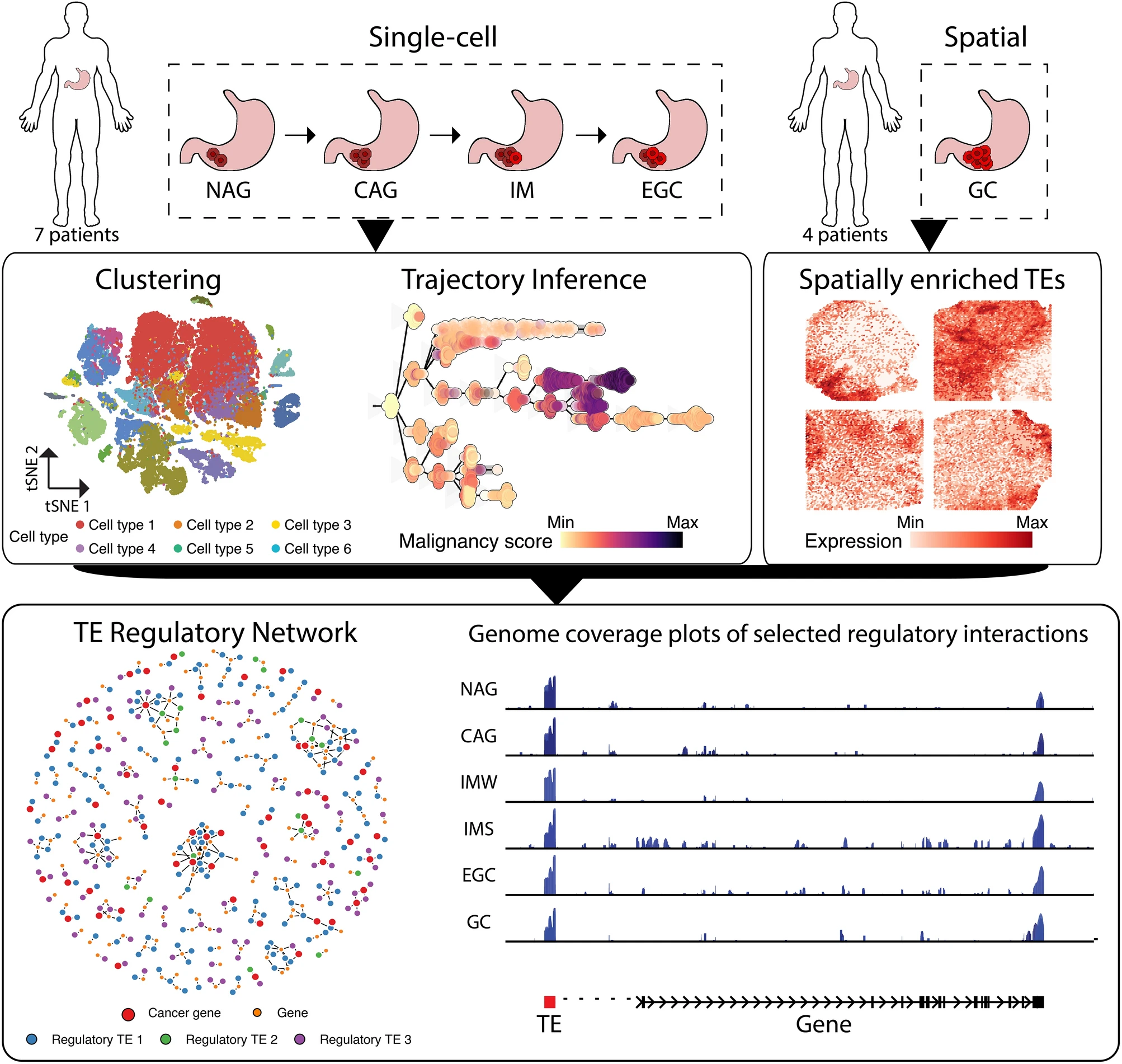 The spatial and cellular portrait of transposable element expression during gastric cancerBraulio Valdebenito-MaturanaScientific Reports, Sep 2024
The spatial and cellular portrait of transposable element expression during gastric cancerBraulio Valdebenito-MaturanaScientific Reports, Sep 2024Gastric Cancer (GC) is a lethal malignancy, with urgent need for the discovery of novel biomarkers for its early detection. I previously showed that Transposable Elements (TEs) become activated in early GC (EGC), suggesting a role in gene expression. Here, I follow-up on that evidence using single-cell data from gastritis to EGC, and show that TEs are expressed and follow the disease progression, with 2,430 of them being cell populations markers. Pseudotemporal trajectory modeling revealed 111 TEs associated with the origination of cancer cells. Analysis of spatial data from GC also confirms TE expression, with 204 TEs being spatially enriched in the tumor regions and the tumor microenvironment, hinting at a role of TEs in tumorigenesis. Finally, a network of TE-mediated gene regulation was modeled, indicating that 2,000 genes could be modulated by TEs, with 500 of them already implicated in cancer. These results suggest that TEs might play a functional role in GC progression, and highlights them as potential biomarker for its early detection.
2022
-
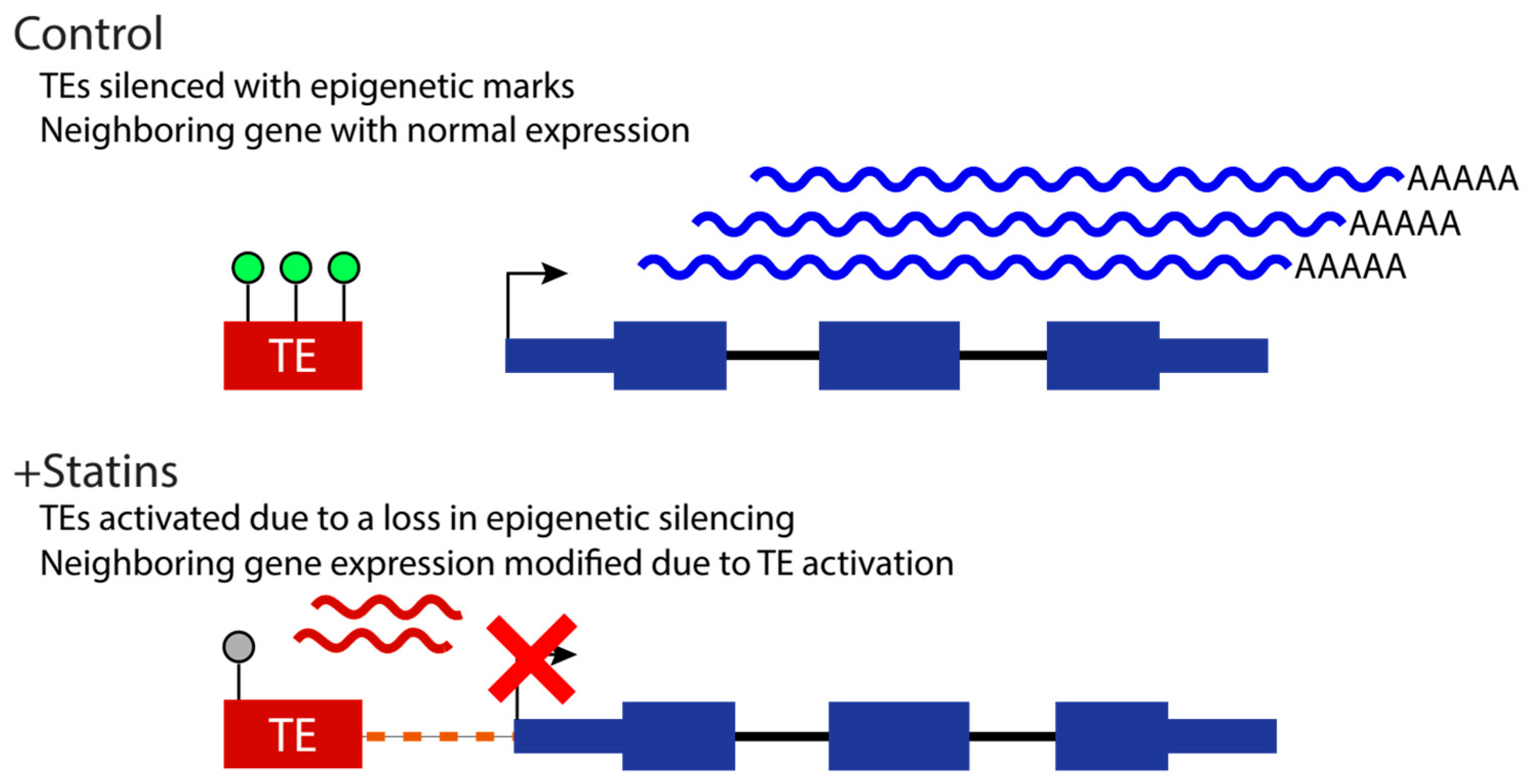 Activation of Transposable Elements in Human Skeletal Muscle Fibers upon Statin Treatment.Braulio Valdebenito-Maturana, Franco Valdebenito-Maturana, Mónica Carrasco, and 2 more authorsInternational journal of molecular sciences, Dec 2022
Activation of Transposable Elements in Human Skeletal Muscle Fibers upon Statin Treatment.Braulio Valdebenito-Maturana, Franco Valdebenito-Maturana, Mónica Carrasco, and 2 more authorsInternational journal of molecular sciences, Dec 2022High cholesterol levels have been linked to a high risk of cardiovascular diseases, and preventative pharmacological care to lower cholesterol levels is critically important. Statins, which are hydroxymethylglutaryl-coenzyme A (HMG-CoA) reductase inhibitors, are drugs used to reduce the endogenous cholesterol synthesis, thus minimizing its pathophysiological effects. Despite the proven benefits, statins therapy is known to cause a number of skeletal muscle disorders, including myalgia, myopathy and myositis. The mechanisms underlying such statin-induced side effects are unknown. Recently, a group of genes and molecular pathways has been described to participate in statin-induced myopathy, caused by either simvastatin or rosuvastatin, although the mechanism by which changes in gene regulation occur was not studied. Transposable Elements (TEs), repetitive elements that move within the genome, are known to play regulatory roles in gene expression; however, their role in statin-induced muscle damage has not been studied. We analyzed the expression of TEs in human skeletal fiber cells treated with either simvastatin or rosuvastatin, as well as their respective controls, and identified TEs that change their expression in response to the treatment. We found that simvastatin resulted in >1000 differentially expressed (DE) TEs, whereas rosuvastatin resulted in only 27 DE TEs. Using network analysis tools, we predicted the impact of the DE TEs on the expression of genes and found that amongst the genes potentially modulated by TEs, there are some previously associated to statin-linked myopathy pathways (e.g., AKT3). Overall, our results indicate that TEs may be a key player in the statin-induced muscle side effects.
- Dysregulated Expression of Transposable Elements in TDP-43M337V Human Motor Neurons That Recapitulate Amyotrophic Lateral Sclerosis In Vitro.Braulio Valdebenito-Maturana, Matias Ignacio Rojas-Tapia, Mónica Carrasco, and 1 more authorInternational journal of molecular sciences, Dec 2022
Amyotrophic lateral sclerosis (ALS) is a disease that progressively annihilates spinal cord motor neurons, causing severe motor decline and death. The disease is divided into familial and sporadic ALS. Mutations in the TAR DNA binding protein 43 (TDP-43) have been involved in the pathological emergence and progression of ALS, although the molecular mechanisms eliciting the disease are unknown. Transposable elements (TEs) and DNA sequences capable of transposing within the genome become dysregulated and transcribed in the presence of TDP-43 mutations. We performed RNA-Seq in human motor neurons (iMNs) derived from induced pluripotent stem cells (iPSCs) from TDP-43 wild-type-iMNs-TDP-43WT-and mutant-iMNs-TDP-43M337V-genotypes at 7 and 14 DIV, and, with state-of-the-art bioinformatic tools, analyzed whether TDP-43M337V alters both gene expression and TE activity. Our results show that TDP-43M337V induced global changes in the gene expression and TEs levels at all in vitro stages studied. Interestingly, many genetic pathways overlapped with that of the TEs activity, suggesting that TEs control the expression of several genes. TEs correlated with genes that played key roles in the extracellular matrix and RNA processing: all the regulatory pathways affected in ALS. Thus, the loss of TE regulation is present in TDP-43 mutations and is a critical determinant of the disease in human motor neurons. Overall, our results support the evidence that indicates TEs are critical regulatory sequences contributing to ALS neurodegeneration.
- SoloTE for improved analysis of transposable elements in single-cell RNA-Seq data using locus-specific expression.Rocío Rodríguez-Quiroz, and Braulio Valdebenito-MaturanaCommunications biology, Oct 2022
Transposable Elements (TEs) contribute to the repetitive fraction in almost every eukaryotic genome known to date, and their transcriptional activation can influence the expression of neighboring genes in healthy and disease states. Single cell RNA-Seq (scRNA-Seq) is a technical advance that allows the study of gene expression on a cell-by-cell basis. Although a current computational approach is available for the single cell analysis of TE expression, it omits their genomic location. Here we show SoloTE, a pipeline that outperforms the previous approach in terms of computational resources and by allowing the inclusion of locus-specific TE activity in scRNA-Seq expression matrixes. We then apply SoloTE to several datasets to reveal the repertoire of TEs that become transcriptionally active in different cell groups, and based on their genomic location, we predict their potential impact on gene expression. As our tool takes as input the resulting files from standard scRNA-Seq processing pipelines, we expect it to be widely adopted in single cell studies to help researchers discover patterns of cellular diversity associated with TE expression.
2021
- Spatially Resolved Expression of Transposable Elements in Disease and Somatic Tissue with SpatialTE.Braulio Valdebenito-Maturana, Cristina Guatimosim, Mónica Alejandra Carrasco, and 1 more authorInternational journal of molecular sciences, Dec 2021
Spatial transcriptomics (ST) is transforming the way we can study gene expression and its regulation through position-specific resolution within tissues. However, as in bulk RNA-Seq, transposable elements (TEs) are not being studied due to their highly repetitive nature. In recent years, TEs have been recognized as important regulators of gene expression, and thus, TE expression analysis in a spatially resolved manner could further help to understand their role in gene regulation within tissues. We present SpatialTE, a tool to analyze TE expression from ST datasets and show its application in somatic and diseased tissues. The results indicate that TEs have spatially regulated expression patterns and that their expression profiles are spatially altered in ALS disease, indicating that TEs might perform differential regulatory functions within tissue organs. We have made SpatialTE publicly available as open-source software under an MIT license.
- Differential regulation of transposable elements (TEs) during the murine submandibular gland development.Braulio Valdebenito-Maturana, Francisca Torres, Mónica Carrasco, and 1 more authorMobile DNA, Oct 2021
The submandibular gland (SG) is a relatively simple organ formed by three cell types: acinar, myoepithelial, and an intricate network of duct-forming epithelial cells, that together fulfills several physiological functions from assisting food digestion to acting as an immune barrier against pathogens. Successful SG organogenesis is the product of highly controlled and orchestrated genetic and transcriptional programs. Mounting evidence links Transposable Elements (TEs), originally thought to be selfish genetic elements, to different aspects of gene regulation in mammalian development and disease. To our knowledge, the role of TEs during murine SG organogenesis has not been studied. Using novel bioinformatic tools and publicly available RNA-Seq datasets, our results indicate that a significant number of genic and intergenic TEs are differentially expressed during the SG development. Furthermore, changes in expression of specific TEs correlated with that of genes involved in cellular division and differentiation, critical aspects for SG maturation. Altogether, we propose that TEs modulate gene networks that operate during SG development.
- Locus-specific analysis of Transposable Elements during the progression of ALS in the SOD1G93A mouse model.Braulio Valdebenito-Maturana, Esteban Arancibia, Gonzalo Riadi, and 2 more authorsPloS one, Oct 2021
Transposable Elements (TEs) are ubiquitous genetic elements with the ability to move within a genome. TEs contribute to a large fraction of the repetitive elements of a genome, and because of their nature, they are not routinely analyzed in RNA-Seq gene expression studies. Amyotrophic Lateral Sclerosis (ALS) is a lethal neurodegenerative disease, and a well-accepted model for its study is the mouse harboring the human SOD1G93A mutant. In this model, landmark stages of the disease can be recapitulated at specific time points, making possible to understand changes in gene expression across time. While there are several works reporting TE activity in ALS models, they have not explored their activity through the disease progression. Moreover, they have done it at the expense of losing their locus of expression. Depending on their genomic location, TEs can regulate genes in cis and in trans, making locus-specific analysis of TEs of importance in order to understand their role in modulating gene expression. Particularly, the locus-specific role of TEs in ALS has not been fully elucidated. In this work, we analyzed publicly available RNA-Seq datasets of the SOD1G93A mouse model, to understand the locus-specific role of TEs. We show that TEs become up-regulated at the early stages of the disease, and via statistical associations, we speculate that they can regulate several genes, which in turn might be contributing to the genetic dysfunction observed in ALS.
- GSER (a Genome Size Estimator using R): a pipeline for quality assessment of sequenced genome libraries through genome size estimation.Braulio Valdebenito-Maturana, and Gonzalo RiadiInterface focus, Jun 2021
The first step in any genome research after obtaining the read data is to perform a due quality control of the sequenced reads. In a de novo genome assembly project, the second step is to estimate two important features, the genome size and ’best k-mer’, to start the assembly tests with different de novo assembly software and its parameters. However, the quality control of the sequenced genome libraries as a whole, instead of focusing on the reads only, is frequently overlooked and realized to be important only when the assembly tests did not render the expected results. We have developed GSER, a Genome Size Estimator using R, a pipeline to evaluate the relationship between k-mers and genome size, as a means for quality assessment of the sequenced genome libraries. GSER generates a set of charts that allow the analyst to evaluate the library datasets before starting the assembly. The script which runs the pipeline can be downloaded from http://www.mobilomics.org/GSER/downloads or http://github.com/mobilomics/GSER.
- Plant HKT Channels: An Updated View on Structure, Function and Gene Regulation.Janin Riedelsberger, Julia K Miller, Braulio Valdebenito-Maturana, and 3 more authorsInternational journal of molecular sciences, Feb 2021
HKT channels are a plant protein family involved in sodium (Na+) and potassium (K+) uptake and Na+-K+ homeostasis. Some HKTs underlie salt tolerance responses in plants, while others provide a mechanism to cope with short-term K+ shortage by allowing increased Na+ uptake under K+ starvation conditions. HKT channels present a functionally versatile family divided into two classes, mainly based on a sequence polymorphism found in the sequences underlying the selectivity filter of the first pore loop. Physiologically, most class I members function as sodium uniporters, and class II members as Na+/K+ symporters. Nevertheless, even within these two classes, there is a high functional diversity that, to date, cannot be explained at the molecular level. The high complexity is also reflected at the regulatory level. HKT expression is modulated at the level of transcription, translation, and functionality of the protein. Here, we summarize and discuss the structure and conservation of the HKT channel family from algae to angiosperms. We also outline the latest findings on gene expression and the regulation of HKT channels.
2018
- TEcandidates: prediction of genomic origin of expressed transposable elements using RNA-seq data.Braulio Valdebenito-Maturana, and Gonzalo RiadiBioinformatics (Oxford, England), Nov 2018
MOTIVATION In recent years, Transposable Elements (TEs) have been related to gene regulation. However, estimating the origin of expression of TEs through RNA-seq is complicated by multi-mapping reads coming from their repetitive sequences. Current approaches that address multi-mapping reads are focused in expression quantification and not in finding the origin of expression. Addressing the genomic origin of expressed TEs could further aid in understanding the role that TEs might have in the cell. RESULTS We have developed a new pipeline called TEcandidates, based on de novo transcriptome assembly to assess the instances of TEs being expressed, along with their location, to include in downstream DE analysis. TEcandidates takes as input the RNA-seq data, the genome sequence and the TE annotation file and returns a list of coordinates of candidate TEs being expressed, the TEs that have been removed and the genome sequence with removed TEs as masked. This masked genome is suited to include TEs in downstream expression analysis, as the ambiguity of reads coming from TEs is significantly reduced in the mapping step of the analysis. AVAILABILITY AND IMPLEMENTATION The script which runs the pipeline can be downloaded at http://www.mobilomics.org/tecandidates/downloads or http://github.com/TEcandidates/TEcandidates. SUPPLEMENTARY INFORMATION Supplementary data are available at Bioinformatics online.
- In silico analysis of the competition between Streptococcus sanguinis and Streptococcus mutans in the dental biofilm.B Valdebenito, P O Tullume-Vergara, W González, and 2 more authorsMolecular oral microbiology, Apr 2018
During dental caries, the dental biofilm modifies the composition of the hundreds of involved bacterial species. Changing environmental conditions influence competition. A pertinent model to exemplify the complex interplay of the microorganisms in the human dental biofilm is the competition between Streptococcus sanguinis and Streptococcus mutans. It has been reported that children and adults harbor greater numbers of S. sanguinis in the oral cavity, associated with caries-free teeth. Conversely, S. mutans is predominant in individuals with a high number of carious lesions. Competition between both microorganisms stems from the production of H2 O2 by S. sanguinis and mutacins, a type of bacteriocins, by S. mutans. There is limited evidence on how S. sanguinis survives its own H2 O2 levels, or if it has other mechanisms that might aid in the competition against S. mutans, nonetheless. We performed a genomic and metabolic pathway comparison, coupled with a comprehensive literature review, to better understand the competition between these two species. Results indicated that S. sanguinis can outcompete S. mutans by the production of an enzyme capable of metabolizing H2 O2 . S. mutans, however, lacks the enzyme and is susceptible to the peroxide from S. sanguinis. In addition, S. sanguinis can generate energy through gluconeogenesis and seems to have evolved different communication mechanisms, indicating that novel proteins may be responsible for intra-species communication.
2017
- Mutantelec: An In Silico mutation simulation platform for comparative electrostatic potential profiling of proteins.Braulio Valdebenito-Maturana, Jose Antonio Reyes-Suarez, Jaime Henriquez, and 4 more authorsJournal of computational chemistry, Mar 2017
The electrostatic potential plays a key role in many biological processes like determining the affinity of a ligand to a given protein target, and they are responsible for the catalytic activity of many enzymes. Understanding the effect that amino acid mutations will have on the electrostatic potential of a protein, will allow a thorough understanding of which residues are the most important in a protein. MutantElec, is a friendly web application for in silico generation of site-directed mutagenesis of proteins and the comparison of electrostatic potential between the wild type protein and the mutant(s), based on the three-dimensional structure of the protein. The effect of the mutation is evaluated using different approach to the traditional surface map. MutantElec provides a graphical display of the results that allows the visualization of changes occurring at close distance from the mutation and thus uncovers the local and global impact of a specific change. \textcopyright 2017 Wiley Periodicals, Inc.
2016
- Cooperation through Competition-Dynamics and Microeconomics of a Minimal Nutrient Trade System in Arbuscular Mycorrhizal Symbiosis.Stephan Schott, Braulio Valdebenito, Daniel Bustos, and 3 more authorsFrontiers in plant science, Mar 2016
In arbuscular mycorrhizal (AM) symbiosis, fungi and plants exchange nutrients (sugars and phosphate, for instance) for reciprocal benefit. Until now it is not clear how this nutrient exchange system works. Here, we used computational cell biology to simulate the dynamics of a network of proton pumps and proton-coupled transporters that are upregulated during AM formation. We show that this minimal network is sufficient to describe accurately and realistically the nutrient trade system. By applying basic principles of microeconomics, we link the biophysics of transmembrane nutrient transport with the ecology of organismic interactions and straightforwardly explain macroscopic scenarios of the relations between plant and AM fungus. This computational cell biology study allows drawing far reaching hypotheses about the mechanism and the regulation of nutrient exchange and proposes that the "cooperation" between plant and fungus can be in fact the result of a competition between both for the same resources in the tiny periarbuscular space. The minimal model presented here may serve as benchmark to evaluate in future the performance of more complex models of AM nutrient exchange. As a first step toward this goal, we included SWEET sugar transporters in the model and show that their co-occurrence with proton-coupled sugar transporters results in a futile carbon cycle at the plant plasma membrane proposing that two different pathways for the same substrate should not be active at the same time.
2015
- K₂p channels in plants and animals.Wendy González, Braulio Valdebenito, Julio Caballero, and 9 more authorsPflugers Archiv : European journal of physiology, May 2015
Two-pore domain potassium (K2P) channels are membrane proteins widely identified in mammals, plants, and other organisms. A functional channel is a dimer with each subunit comprising two pore-forming loops and four transmembrane domains. The genome of the model plant Arabidopsis thaliana harbors five genes coding for K2P channels. Homologs of Arabidopsis K2P channels have been found in all higher plants sequenced so far. As with the K2P channels in mammals, plant K2P channels are targets of external and internal stimuli, which fine-tune the electrical properties of the membrane for specialized transport and/or signaling tasks. Plant K2P channels are modulated by signaling molecules such as intracellular H(+) and calcium and physical factors like temperature and pressure. In this review, we ask the following: What are the similarities and differences between K2P channels in plants and animals in terms of their physiology? What is the nature of the last common ancestor (LCA) of these two groups of proteins? To answer these questions, we present physiological, structural, and phylogenetic evidence that discards the hypothesis proposing that the duplication and fusion that gave rise to the K2P channels occurred in a prokaryote LCA. Conversely, we argue that the K2P LCA was most likely a eukaryote organism. Consideration of plant and animal K2P channels in the same study is novel and likely to stimulate further exchange of ideas between students of these fields.