On the importance of assessing the results of computational tools with ground truth data
This is a supplementary material for my upcoming book chapter “Transposable Element analysis in OMICS data”
PRELIMINARIES
In a previous post, I briefly mentioned how the tool SQuIRE, which can be used for locus-specific measurements of Transposable Element (TE) expression from RNA-Seq data, can results in false positives. Here, I will expand on that, by showing a fully reproducible example of this. This post is not intended to discredit SQuIRE, but rather to show that for any computational tool we always need to think on ways to test their results with ground truth data, that is, with inputs that we know for a fact that should result in a specific output. In this line, here I designed a simple experiment to show how this can be achieved.
WHAT YOU WILL NEED
- A STAR-indexed genome
- Experiment sequences
- ART read simulator
- SQuIRE
- IGV
STEP 1 - GENOME INDEX
The first step is to get the genome FASTA file and GTF gene annotation file:
wget http://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/hg38.fa.gz
wget http://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/genes/hg38.ncbiRefSeq.gtf.gz
gzip -d hg38.fa.gz
gzip -d hg38.ncbiRefSeq.gtf.gz
We can then build the index with STAR as follows:
STAR --runMode genomeGenerate --runThreadN 21 --genomeDir hg38_STAR_INDEX --genomeFastaFiles hg38.fa --sjdbGTFfile hg38.ncbiRefSeq.gtf
STEP 2 - EXPERIMENT SEQUENCES
For this simple experiment, in the experiment_sequences.fa file, we only have two sequences:
grep "^>" experiment_sequences.fa
## >chr4|4013499|4019703|L1PA4:L1:LINE|5.8|-::chr4:4013499-4019703(-)
## >NM_020364.4 Homo sapiens deleted in azoospermia 3 (DAZ3), mRNA
One is a sequence from a TE located in chromosome 4, while the other is from the protein-coding sequence of the DAZ3 gene located in chromosome Y.
STEP 3 - READ SIMULATION
We can get the ART read simulator from from its official website:
wget https://www.niehs.nih.gov/sites/default/files/2024-02/artbinmountrainier2016.06.05linux64.tgz
tar -xvf artbinmountrainier2016.06.05linux64.tgz
To perform read simulation, we set then -ss HS25 for single-end layout, -l 150 for read length of 150 bp, -f 30 to simulate reads representing 30X coverage of the sequences in -i experiment_sequences.fa, and -o experiment_sequences as output prefix.
art_bin_MountRainier/art_illumina -ss HS25 -i experiment_sequences.fa -l 150 -f 30 -o experiment_sequences
This will give us the experiment_sequences.fq file, which has the FASTQ simulated reads to map against the human genome.
STEP 4 - SETTING UP SQUIRE
The installation of SQuIRE is a relatively seamless process if you already have Conda. Their repository is well documented and provides instruction to install the tool from scratch, including setting up Conda. You can check their instructions here
Once installed, we need to check that the Count tool is available:
squire Count -h
usage: squire Count [-h] [-m <folder>] [-c <folder>] [-o <folder>]
[-t <folder>] [-f <folder>] -r <int> [-n <str>]
[-b <build>] [-p <int>] [-s <int>] [-e EM] [-v]
Arguments:
-h, --help show this help message and exit
-m <folder>, --map_folder <folder>
Folder location of outputs from SQuIRE Map (optional,
default = 'squire_map')
-c <folder>, --clean_folder <folder>
Folder location of outputs from SQuIRE Clean
(optional, default = 'squire_clean')
-o <folder>, --count_folder <folder>
Destination folder for output files(optional, default
= 'squire_count')
-t <folder>, --tempfolder <folder>
Folder for tempfiles (optional; default='count_folder')
-f <folder>, --fetch_folder <folder>
Folder location of outputs from SQuIRE Fetch
(optional, default = 'squire_fetch)'
-r <int>, --read_length <int>
Read length (if trim3 selected, after trimming;
required).
-n <str>, --name <str>
Common basename for input files (required if more than
one bam file in map_folder)
-b <build>, --build <build>
UCSC designation for genome build, eg. 'hg38'
(required if more than 1 build in clean_folder)
-p <int>, --pthreads <int>
Launch <int> parallel threads(optional; default='1')
-s <int>, --strandedness <int>
'0' if unstranded eg Standard Illumina, 1 if first-
strand eg Illumina Truseq, dUTP, NSR, NNSR, 2 if
second-strand, eg Ligation, Standard SOLiD
(optional,default=0)
-e EM, --EM EM Run estimation-maximization on TE counts given number
of times (optional, specify 0 if no EM desired;
default=auto)
-v, --verbosity Want messages and runtime printed to stderr (optional;
default=False)
We now need to run these commands to prepare some additional files and directories required for the quantification of TE expression:
squire Fetch -b hg38 -c -g -r -v
squire Clean -r squire_fetch/hg38_rmsk.txt -v
Now, we should have the directories squire_fetch and squire_clean:
tree squire_fetch
tree squire_clean
## squire_fetch
## ├── hg38chromFa.tar.gz
## ├── hg38_chromInfo.txt
## ├── hg38_refGene.bed
## ├── hg38_refGene.genepred
## ├── hg38_refGene.gtf
## └── hg38_rmsk.txt
##
## 0 directories, 6 files
## squire_clean
## ├── hg38_all.bed
## └── hg38_all_copies.txt
##
## 0 directories, 2 files
STEP 5 - MAPPING
Now we are all set!
We can map our simulated sequences experiment_sequences.fq against the genome index, using the same options that squire Map uses
STAR --runThreadN 1 --clip3pNbases 0 --outFilterMultimapNmax 100 --winAnchorMultimapNmax 100 --genomeDir hg38_STAR_INDEX --readFilesIn experiment_sequences.fq --outFileNamePrefix squire_map/experiment_alignment_ --outSAMtype BAM SortedByCoordinate --outSAMattributes All --outSAMstrandField intronMotif --outSAMattrIHstart 0 --sjdbGTFfile hg38.ncbiRefSeq.gtf --twopassMode Basic
tree squire_map
## squire_map
## ├── experiment_alignment_Aligned.sortedByCoord.out.bam
## ├── experiment_alignment_Aligned.sortedByCoord.out.bam.bai
## ├── experiment_alignment_Log.final.out
## ├── experiment_alignment_Log.out
## ├── experiment_alignment_Log.progress.out
## ├── experiment_alignment_SJ.out.tab
## ├── experiment_alignment__STARgenome
## │ ├── exonGeTrInfo.tab
## │ ├── exonInfo.tab
## │ ├── geneInfo.tab
## │ ├── sjdbInfo.txt
## │ ├── sjdbList.fromGTF.out.tab
## │ ├── sjdbList.out.tab
## │ └── transcriptInfo.tab
## └── experiment_alignment__STARpass1
## ├── Log.final.out
## └── SJ.out.tab
##
## 2 directories, 15 files
STEP 6 - TE QUANTIFICATION AND VALIDATION
With the mapping result, we can run the Count quantification tool:
squire Count -m squire_map/ -r 150 -c squire_clean/ -v
tree squire_count
## squire_count
## ├── experiment_alignment_Aligned.sortedByCoord.out_abund.txt
## ├── experiment_alignment_Aligned.sortedByCoord.out.gtf
## ├── experiment_alignment_Aligned.sortedByCoord.out_refGenecounts.txt
## ├── experiment_alignment_Aligned.sortedByCoord.out_subFcounts.txt
## └── experiment_alignment_Aligned.sortedByCoord.out_TEcounts.txt
##
## 0 directories, 5 files
Of these files, we are interested in experiment_alignment_Aligned.sortedByCoord.out_TEcounts.txt, which provides the quantification of TEs. We can check the most important columns:
awk 'BEGIN{FS=OFS="\t"}{print $1,$2,$3,$4,$15,$16,$17}' squire_count/experiment_alignment_Aligned.sortedByCoord.out_TEcounts.txt
## tx_chr tx_start tx_stop TE_ID uniq_counts tot_counts tot_reads
## chr4 4013502 4019680 chr4|4013498|4019703|L1PA4:L1:LINE|58|- 1118 1183.90 1216
## chr8 96955960 96956120 chr8|96955434|96956163|L1PA4:L1:LINE|47|+ 0 4.31 6
## chr10 22061677 22061836 chr10|22060724|22066859|L1PA5:L1:LINE|53|+ 0 4.20 6
## chr4 9617631 9617845 chr4|9617608|9619322|L1PA4:L1:LINE|86|+ 0 5.29 15
## chr10 82344563 82344736 chr10|82344062|82350086|L1PA2:L1:LINE|25|- 0 3.79 12
## chr3 112659647 112659820 chr3|112659127|112665164|L1PA3:L1:LINE|29|- 0 3.79 12
## chr5 62656507 62656735 chr5|62655618|62660602|L1P1:L1:LINE|57|+ 0 3.34 14
## chr5 93766091 93766242 chr5|93761070|93767199|L1PA4:L1:LINE|55|+ 0 1.73 2
## chr2 133466941 133467107 chr2|133465007|133471038|L1PA2:L1:LINE|29|- 0 1.53 5
## chr20 29366629 29366779 chr20|29366059|29372220|L1PA4:L1:LINE|54|- 1 1.00 1
## chr5 114891825 114891975 chr5|114891256|114895798|L1PA4:L1:LINE|49|- 1 1.00 1
## chrX 97578708 97578858 chrX|97578170|97584188|L1PA3:L1:LINE|25|- 1 1.00 1
## chrX 75693308 75693519 chrX|75690170|75694093|L1PA4:L1:LINE|58|- 0 1.24 15
## chr3 115315428 115315678 chr3|115312305|115318470|L1PA3:L1:LINE|34|- 0 1.45 22
## chr3 121455034 121455284 chr3|121452690|121458395|L1PA3:L1:LINE|39|+ 0 1.45 22
## chr12 29982525 29982675 chr12|29981945|29982867|L1PA4:L1:LINE|29|- 0 0.50 1
## chr1 48118716 48118866 chr1|48118135|48120412|L1PA4:L1:LINE|31|- 0 0.50 1
## chrY 24773569 24784208 chrY|24776006|24782046|L1PA2:L1:LINE|31|+ 15 15.00 15
Wait a minute!!. So, from our experiment design, we should only expect TE expression at chromosome 4, specifically at chr4:4013499-4019703. We indeed see this on the first row, but there is expression detected at other chromosomes:
awk 'BEGIN{FS=OFS="\t"}{print $1}' squire_count/experiment_alignment_Aligned.sortedByCoord.out_TEcounts.txt|sort|uniq -c
## 1 chr1
## 2 chr10
## 1 chr12
## 1 chr2
## 1 chr20
## 3 chr3
## 2 chr4
## 3 chr5
## 1 chr8
## 2 chrX
## 1 chrY
## 1 tx_chr
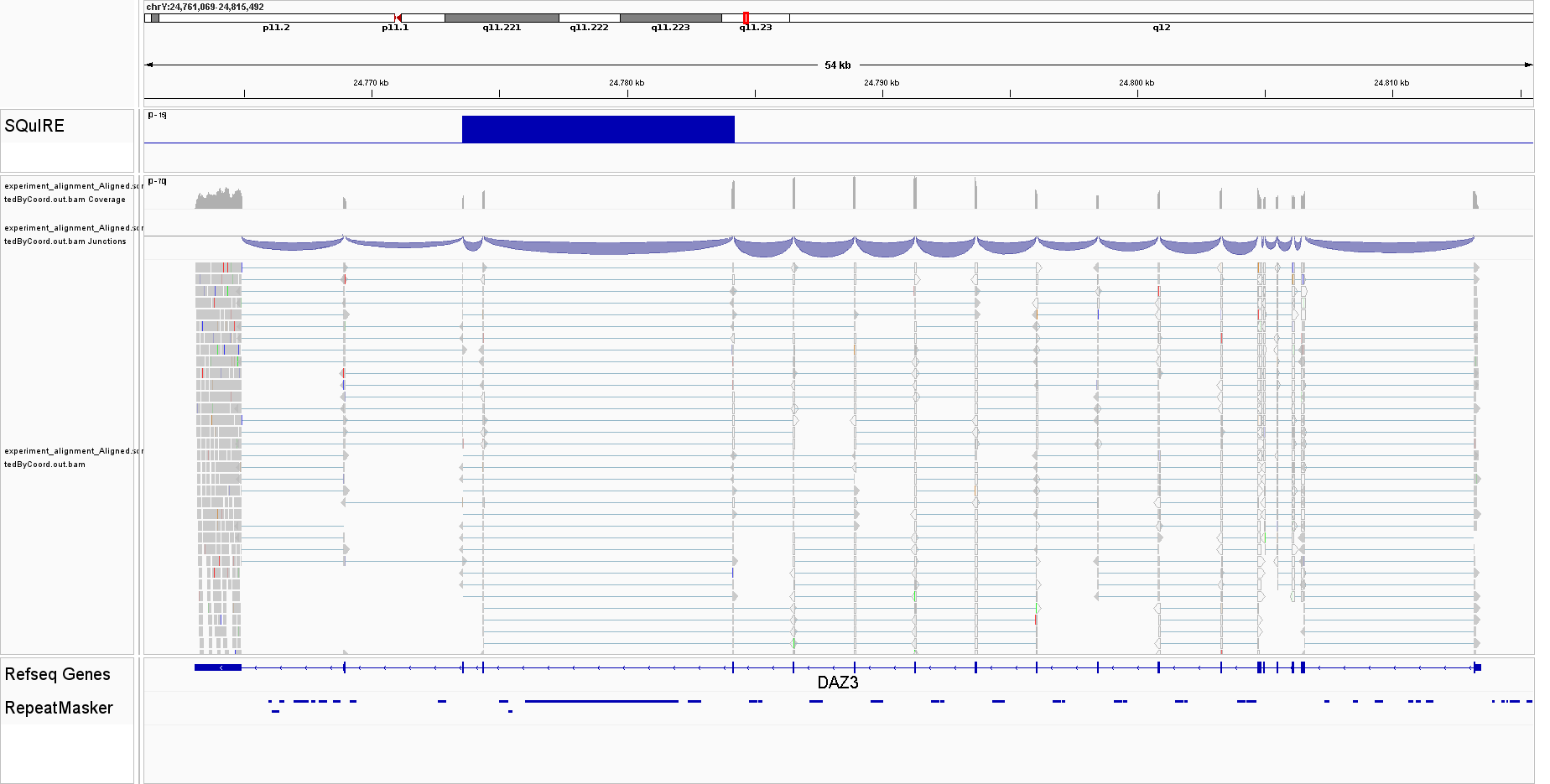
We see a result in chromosome Y, at locations 24773569 to 24784208:
awk 'BEGIN{FS=OFS="\t"}{print $1,$2,$3,$4}' squire_count/experiment_alignment_Aligned.sortedByCoord.out_TEcounts.txt|grep "chrY"
## chrY 24773569 24784208 chrY|24776006|24782046|L1PA2:L1:LINE|31|+
This regions overlap with the location of DAZ3, approximately at chrY:24761069-24815492, which is the gene from which we also simulated sequences.
We can now inspect these results in IGV. First, we create a bedgraph representation of our BAM file using the bamCoverage tool from deepTools:
bamCoverage --bam squire_map/experiment_alignment_Aligned.sortedByCoord.out.bam --outFileName experiment.bedgraph --outFileFormat bedgraph --binSize 5 --region chrY:24761069:24815492
A dummy representation of SQuIRE’s results as bedgraph can be created as follows:
echo -e "chrY\t24773569\t24784208\t15" > squire.bedgraph
Those 2 files can be seamlessly loaded in IGV, and we will be able to see this:
If proficient with ggCoverage, we can create a similar representation of the IGV result, as a fully vectorial image:
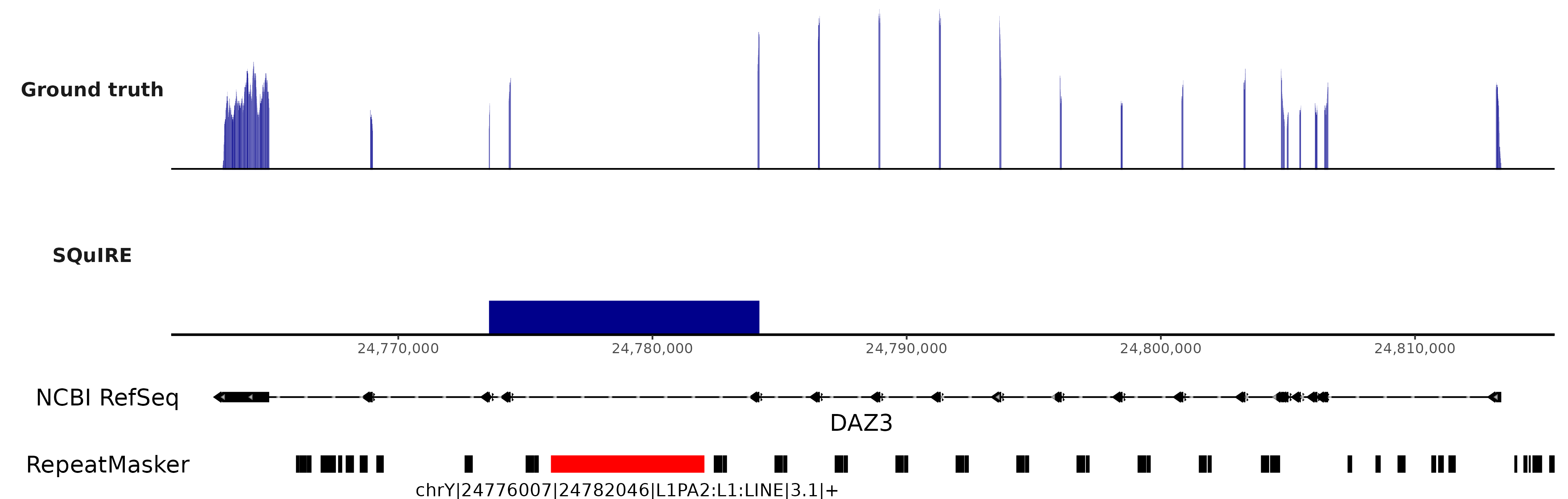
I found that SQuIRE over-estimates intronic TE expression due to indiscriminate use of BEDtools. When there are spliced alignments, BEDtools by default will convert the interval in the SAM/BAM file to start position plus length of CIGAR string. As an example, for a CIGAR string like 20M3000N80N, and alignment position at 1000000, it will create an interval of length 20+3000+80 = 3100, and end position at 1003100. We need to either use bedtools bamtobed with the -split option to create an intermediate file (which we can further check that correctly has split intervals), or use the -split option in bedtools intersect so it does not artifically creates intervals longer than the sequencing reads.
This is why whenever I use a tool, I design what I call “ground truth” experiments, in which with a simple test dataset, I can accurately tell if the tool does what it is supposed to do. Although it is very exciting that nowadays bioinformatics tool become more readily available, we should always exercise caution in their use, as this can result in arriving at biological conclusions that might not be entirely true. For example, using SQuIRE, a previous article made a point of “intron retention” affecting TE quantification. From my experience, and the experiment shown here, I could argue that there migh t be over-estimation of intronic TE expression, but that does not correspond to actual “intron retention”.
Enjoy Reading This Article?
Here are some more articles you might like to read next: